Everything you need in a genome browser
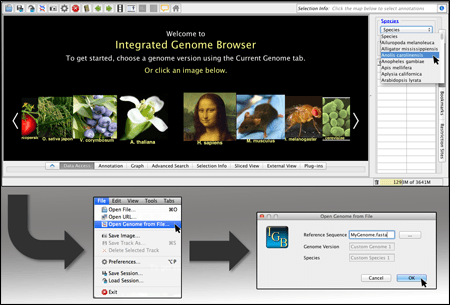
The Integrated Genome Browser (IGB) is an open-source tool for visualizing genome-scale datasets.
IGB is designed for both biologists and bioinformaticians, and is used by thousands of researchers
at hundreds of universities around the world. Find out how IGB can help push your research forward with its many features,
ease of use, and wealth of support documentation.
Need help getting started with IGB? Contact us for help or to schedule a tutorial.
Display your own data
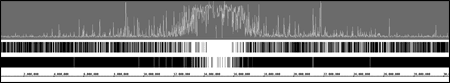
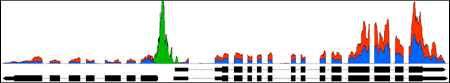
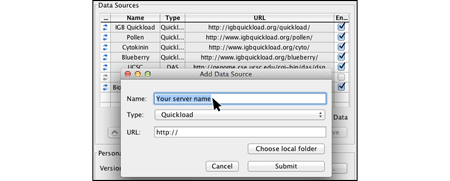
View data from aligned sequences, annotations, and more (BAM, BED, GFF3). Overlap multiple datasets and customize each track's color. Or make a heatmap of the data.
- Accepted file formats
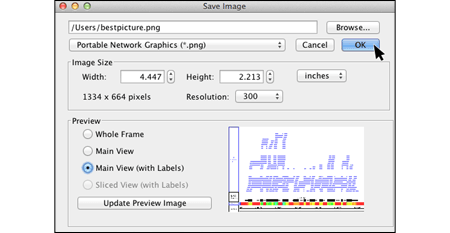
- Set up a heatmap


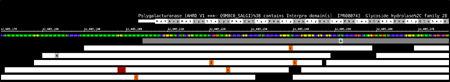
View and interact
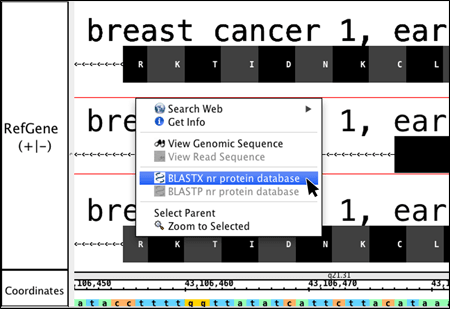
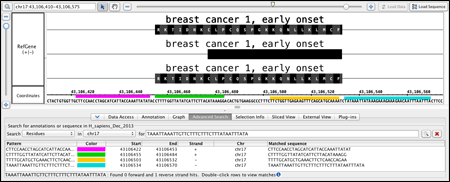
Fly through your data using IGB's fast, animated zooming and rapid panning. Interact with your data by selecting or right-clicking features.
- Moving in IGB
- Data interaction
Advanced Features
Develop and enable Plug-ins
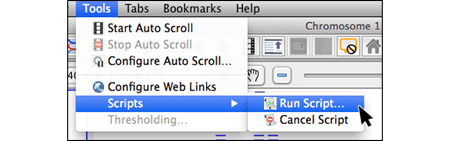
Select and install Plug-ins through the IGB App Manager to add additional functionality. You can also make your own plug-ins/apps and share them with the IGB community.
- Installing plug-ins through App Manager
- Creating new plug-ins/apps
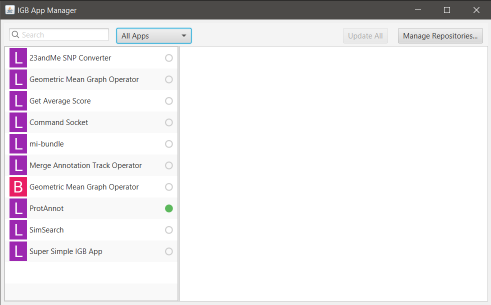
More Information
For more information about IGB capabilities, visit the IGB User's Guide or the IGB Developer's Guide.